PlantAligDB: A database of curated nucleotide sequence alignments for plants
The PlantAligDB is a comprehensive on-line resource of curated nucleotide sequence alignments for plant research. The website provides a complete, quality checked and regularly updated collection of alignments that can be used in taxonomic, molecular genetics, phylogenetic and evolutionary studies.

|
> Explore different genomic regions. > Search for a plant family. > BLAST search against all nucleotide sequences. > Download curated alignments. > Upload your datasets. > Retrieve genetic diversity indices. > Visualize and edit phylogenetic trees. |
|
Database content
|
Nº of different families: 223 Nº of sequences: 66052 Nº of alignments: 514 |
Families with more alignments: Asteraceae Orchidaceae Poaceae |
Largest sequence alignment: Fabaceae (2599 trnL GH sequences) |
Our nucleotide sequence alignments were carefully selected to represent different genomic regions and plant families with at least 10 species.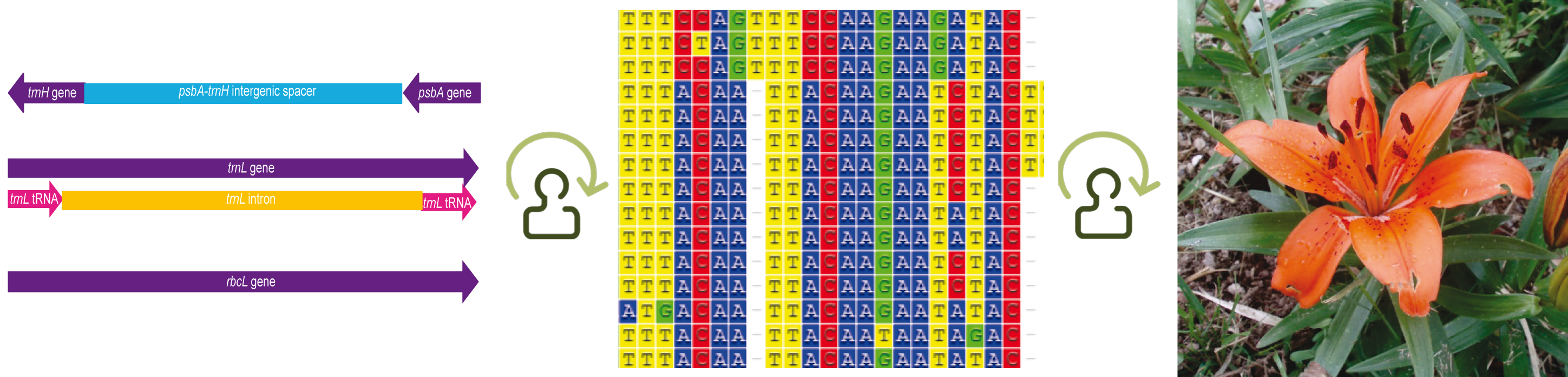 |
| PlantAligDB current statistics (updated March, 2017): | ||||
| Genomic region | Genome | Type | Nº of sequences | Nº of alignments/families |
|---|---|---|---|---|
| atpF-atpH | cpDNA | Inter-genic spacer | 1025 | 31 |
| psbA-trnH | cpDNA | Inter-genic spacer | 4852 | 79 |
| trnL CD | cpDNA | Intron | 2527 | 44 |
| trnL GH | cpDNA | Intron | 34674 | 173 |
| rbcL | cpDNA | Protein-coding gene | 1748 | 39 |
| matK | cpDNA | Protein-coding gene | 11341 | 113 |
| ITS | nuDNA | Transcribed spacers and 5.8S gene | 9885 | 35 |
Taxonomic groups
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Recent news-Last update:09-07-2018.
|
How to cite"A web-based platform of nucleotide sequence alignments of plants"
|